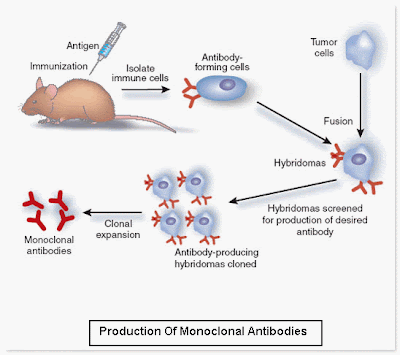
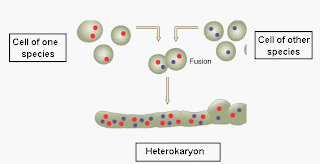
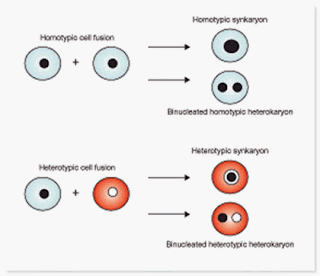
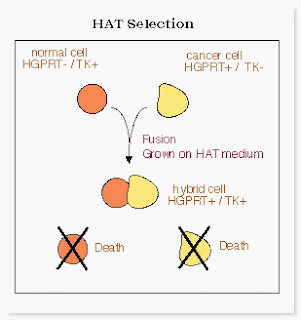
The term molecular biology was first used in 1945 by William Asbury who was referring to the study of the chemical and physical structure of biological macromolecules.
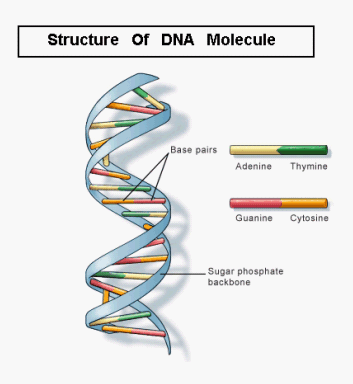
The roots of molecular biology were established in 1953 when an Englishman, Francis Crick and young American, James Watson working at Medical Research Council Unit, Cavendish Laboratory, Cambridge, proposed a double helical model for the structure of DNA (deoxyribonucleic acid) molecule.
Earlier, discoveries made by Griffith (1928), Avery, Macleod, and McCarthy (1944) had clearly revealed that the DNA was the chemical bearer of genetic information of certain microorganisms (bacteria, bacteriophage, etc).
The discoveries of these scientists brought a great revolution and numerous research works, of many scientists all over the world, also confirmed that DNA was the genetic material in plants, animals, and other microorganisms.
All these studies and discoveries has emerged the realization that the basic chemical organization and the metabolic process of all living things are remarkably similar, despite their morphological diversity.
In most organism the phenotype or the body structure and function ultimately depend on the structural and functional proteins or polypeptides. The synthesis of polypeptides is regulated and governed by self-duplicating genes which are born within molecules of DNA.
The genetic information for polypeptide synthesis is initially dictated by the disposition of nitrogen bases in DNA molecule and is copied down by the process of transcription. In transcription multiple copies of an individual gene is synthesized.
These copies are molecules of RNA (ribonucleic acid) such as ribosomal RNA (r-RNA) , messenger RNA (m-RNA) , and transfer RNA (t-RNA) . These RNA molecules lead to the synthesis of polypeptide chain, and this process is called translation.
In translation the genetic message encoded in messenger RNA molecule is translated into linear sequence of amino acids in a polypeptide. This polypeptide in its turn determines the phenotype of the organism.
Background of Molecular Biology
In 1928 F. Griffith discovered transformation in bacteria.
In 1934 M. Schlesinger demonstrated that bacteriophage is composed of DNA and proteins.
In 1944 Avery, Macleod, and McCarthy first reported that DNA and not protein is the hereditary chemical.
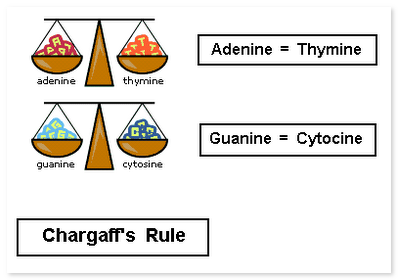
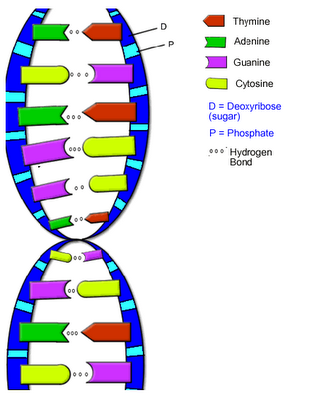
In 1950 Chargaff demonstrated that in DNA the number of adenine and thymine groups is always equal and so are the numbers of guanine and cytosine groups. This is called as Chargaff's Rule.
In 1953 J.D. Watson and F.H.C. Crick proposed the helical structure of DNA.
In 1957 H. Fraenkel-Conart and B. Singer confirmed that RNA is the genetic material of some viruses.
In 1958 G. Beadle and E. Tatum worked on the biochemical genetics of fungus.
In 1959 S. Ochoa and A. Kornberg received Nobel Prize for artificial synthesis of nucleic acid.
In 1964 R.W. Holly gave detailed structure of alanyl tRNA from yeast.
In 1965 F.H.C. Crick proposed the Wobble Hypothesis for anticodons of tRNA. Another scientist F. Jacob and J. Monad received Nobel Prize for the protein synthesis mechanism in virus.
In 1968 R.W. Holly, H.G. Khorana and M.W.Neirenberg got Nobel Prize for deciphering the genetic code.
In 1969 H. Temin and D. Baltimore demonstrated the synthesis of DNA on RNA template tumor viruses. Both were awarded Nobel Prize 1975 for the discovery of enzyme called reverse transcriptase, which is present in the core of virusparticle.
In 1973 S.H. Kim suggested a three dimensional structure (L-shaped Model) of tRNA.
In 1975 E.M. Southern developed method called Southern Blotting Technique for analyzing the related genes in a DNA restriction fragment.
In 1977 P.A. Sharp and R.J. Roberts discovered split genes of adenovirus.
In 1978 W. Gilbert first of all used the term exon and intron (for split genes).
In 1979 Khorana reported completion of the total synthesis of a biologically functional gene. Alwini developed Northern Blotting Technique for m-RNA and Towbin et al. developed the Western Blotting Technique for proteins.
In 1982 A. Klug was awarded Nobel Prize for providing three-dimensional structure of tRNA.
In 1985 K. Mullis discovered polymerase chain reaction (PCR) which is widely exploited in gene cloning for genetic engineering.
In 1984, 86 A. Jaffrey discovered the techniques of DNA fingerprinting.
In 1989 T. Cech and S. Altman were awarded Nobel Prize for showing enzymatic roles of some RNA molecules like ribozymes.
In 1991 Dr. Lalji Singh developed a new technique of DNA fingerprinting by using BKM-DNA probe (BKM=banded krait minor satellite).
Structure of DNA
In prokaryotes DNA is without any associated protein, but in eukaryotes it is combined with histone protein to form nucleoprotein.
DNA is mainly present in chromosomes and also reported in cytoplasmic organelles like mitochondria and chloroplasts.
Rosalind Franklin first of all studied the structure of the DNA by conducting an experiment using X-rays. Her experiment showed that DNA has a helical structure and which was repeated after a certain interval.
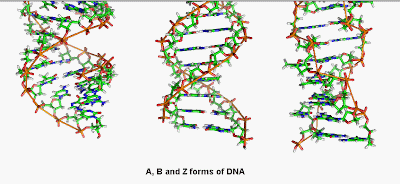
DNA can exist in 5 forms: A, B, C, D, E, and Z. Sugar puckering is the most important feature to distinguishing the DNA forms.
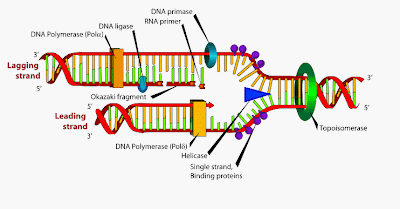
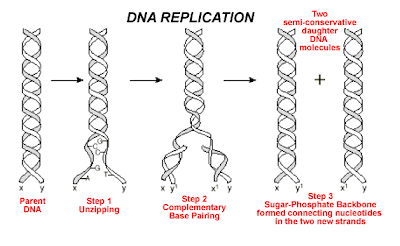
Unwinding: - The old strand that makes up the parent DNA molecule is unwound and unzipped (weak hydrogen bonds between the pared bases are broken). The hydrogen bonds between the molecules are broken with the help of helicase enzyme.
Complementary base pairing: - With the help of enzyme DNA polymerase new complementary nucleotides (that are always present in the nucleus) are positioned adjacent to each other opposite to the parent DNA template.
Joining:-This step also requires DNA polymerase enzyme for joining the complementary nucleotides. Each daughter molecule contains an old and a new strand. replication of DNA strand has an origin point at which the replication is initiated. It may also have a terminus point where the replication of DNA is terminated. A ' Y ' shaped structure is formed at the point of replication which is called as "replication fork ".
These bases are connected with the deoxyribose sugar molecules. The two strands intertwined in a clockwise direction and run in opposite directions. The distance between two adjacent base pair is 3.5 Armstrong and each turn is completed after 10 base pairs. Thus the total distance of one turn is 34 Armstrong.
Different forms of DNA
The above described Watson and Crick model of a DNA molecules contains the right handed helical coiling and has been called B-DNA, which is biologically important form of a DNA found in most of the living organisms.
A-form and C-form differ from the B-form in number of monomers per turn or spacing of residues along helical strands.
D-form and E-form of DNA are found in some organisms and they lack in guanine (nitrogenous base).
Z-DNA is a left handed DNA molecule and one complete helix contains 12 base pairs. The angle of rotation in Z-DNA is 60 degree while that of B-DNA is 36 degree.
Replication of DNA
DNA exhibits autocatalytic function according to which the synthesis of DNA (duplication) is under the control of DNA molecule itself.
In the replication process the parent DNA molecule unwinds and unzips. Then each of the old strands serves as the template for the new strands. Each daughter DNA molecule receives one parental strand and a newly synthesized strand. This type of DNA replication is commonly called as semi-conservative replication, because here each daughter DNA molecule receives one parental strand.
DNA replication requires following three steps:
Replication of DNA may be unidirectional or bidirectional. During DNA replication, one nucleotide is joined with another. Each nucleotide already has a phosphate group at the 5' carbon atom and it is joined to 3' carbon atom of the sugar molecule.
Thus the synthesis of DNA molecule takes place in the 5'->3' direction with the help of DNA polymerase enzyme. But this causes a problem at the replication fork where only one of the new strands run in the 5'->3' direction ( the template for this strand runs in the 3'->5' direction). This strand is called as leading strand.
The template for the other strand runs in the 5'->3' direction, but DNA synthesis could only take place in 5'->3' direction. Thus, this poses a problem and due to this reason synthesis has to begin in the replication fork.
Replication of the 5'->3' parental strand begins as soon as the DNA molecule unwinds and unzips replication of this strand is discontinuous. The replication of this strand results in segment called Okazaki fragments.
Discontinuous replication takes more time than continuous replication therefore the new strand in this case is called the lagging strand.